local extensions (local.gcl)
ASCII
To export an x,y,e array of your workspace use the routine "ascii_export", it takes two parameters:
ascii_export work_space "file name"
work_space=ascii_import("file name","type")
in3
To save a work space to an intermediate genie, in3, file you type
put/new work_space file="name.in3"
work_space=get(3,"name.in3")
graphics
You can change the colour of your next plot by the
commands red green blue and white. A small plot window can be
opened with the command fonster.
dae
To display the dae you type assign "dae" where you have to type
the quotation marks since dae is a string. You can, however type
only assign and the default value is to set your input to the dae
correctL
You can correct old runs with wrong angle and flight path with this command. Either you have a detector.dat file you want to use to replace both angle and flight-path. It is also possible to have a genie intermediate file that hold shift of each detector. The output file will have the extension caw instead of raw and the commands should look like:
correctL/all run_number "detector.dat"
correctL run_number "dL.in3"
5.2 Diffraction
OSIRIS routines (osd.gcl)
There are to different ways of doing reduction for the diffraction data.
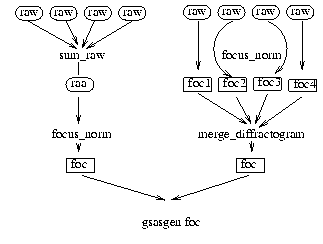
sum_raw
The program takes at least two parameters
sum_raw first_run_number last_run_number [%low %high]
The routine unwrap is used to unwrap the monitor. This is
done by searching for the minimum value in the spectra and then moving
everything from the right of that point to the left side of the spectra.
focus_norm
This routine reads "raw", "sav" or "raa" files. The units are changed to wavelength the monitor is corrected for efficiency (see Monitor) and, detector efficiency corrections are performed Units are changed to d-spacing and the spectra are focused and normalised with the monitor. The spectra are focused in d-space. This routine takes three parameters.
work_space=focus_norm(run_number,first_detector,last_detector,"ext")
focus_norm_bank (fnb)
Same as focus_norm but takes slightly different parameters
work_space=focus_norm_bank(run_number,"bank","ext")
bank=all and ext=raw so to focus all detectors from
a raw file you need only to type
work_space=fnb(run_number)
merge_diffractogram
If you run focus_norm on your raw files you will have to merge the the workspaces into which you have focused your raw data.
work_space=merge_diffractogram(f1,f2,f3...)
ccslgen
A workspace can be exported to ccsl format (three columns x,y,e) with the command
ccslgen work_space
ISIS routines
gsasgen
A workspace can be exported to gsas format with the command
gsasgen work_space
You should choose bank 1 for "all" detectors and bank 2 for the
"high" resolution option.
Next Previous Contents